Kim, D., Kesavan, R., Ryu, K., Dey, T., Marckx, A., Menezes, C., Praharaj, P.P., Morley, S., Ko, B., Soflaee, M.H., Tom, H.J., Brown, H., Vu, H.S., Tso, S.C., Brautigam, C.A., Lemoff, A., Mettlen, M., Mishra, P., Cai, F., Allen, D.K., and G. Hoxhaj. (2025). Mitochondrial NADPH fuels mitochondrial fatty acid synthesis and lipoylation to power oxidative metabolism. Nature Cell Biology (online ahead of print). (PubMed)
Praharaj, P.P.,* Li, Y.,* Mary, C.,* Soflaee, M.H., Ryu, K., Kim, D., Tran, D.H., Dey, T., Tom, H.J., Rion, H., Gelin, M., Lemoff, A., Zacharias, L.G., Patricio, J.S., Mathews, T.P., Chen, Z., Lionne, C., Hoxhaj, G.,# and G. Labesse.# (2025). Cryo-EM structure and regulation of human NAD kinase. Science Advances 11:eads2664. (PubMed)
Tran, D.H.,* Kim, D.,* Kesavan, R.,* Brown, H., Dey, T., Soflaee, M.H., Vu, H.S., Tasdogan, A., Guo, J., Bezwada, D., Al Saad, H., Cai, F., Solmonson, A., Rion, H., Chabatya, R., Merchant, S., Manales, N.J., Tcheuyap, V.T., Mulkey, M., Mathews, T.P., Brugarolas, J., Morrison, S.J., Zhu, H., DeBerardinis, R.J., and G. Hoxhaj. (2024). De novo and salvage purine synthesis pathways across tissues and tumors. Cell 187, 3602-18. (PubMed)
Mary, C.,* Soflaee, M.H.,* Kesavan, R., Gelin, M., Brown, H., Zacharias, L.G., Mathews, T.P., Lemoff, A., Leonne, C., Labesse, G.,# and G. Hoxhaj.# (2022). Crystal structure of human NADK2 reveals a dimeric organization and active site occlusion by lysine acetylation. Molecular Cell 82, 3299-3311. (PubMed)
Soflaee, M.H.,* Kesavan, R.,* Sahu, U.,* Tasdogan, A., Villa, E., Djabari, Z., Cai, F., Tran, D.H., Vu, H.S., Ali, E.S., Rion, H., O’Hara, B.P., Kelekar, S., Hallett, J.H., Martin, M., Mathews, T.P., Gao, P., Asara, J.M., Manning, B.D.#, Ben-Sahra, I.,# and G. Hoxhaj.# (2022). Purine nucleotide depletion prompts cell migration by stimulating the serine synthesis pathway. Nature Communications 13, 2698. (PubMed)
Tran, D.H.,* Kesavan, R.,* Rion, H., Soflaee, M.H., Solmonson, A., Bezwada, D., Vu, H.S., Cai, F., Phillips, J.A.3rd, DeBerardinis, R.J., and G. Hoxhaj. (2021). Mitochondrial NADP+ is essential for proline biosynthesis during cell growth. Nature Metabolism 3, 571-585. (PubMed)
Hoxhaj, G.,# and B.D. Manning.# (2020). The PI3K-AKT network at the interface of oncogenic signalling and cancer metabolism. Nature Reviews Cancer 116, 2539-2544. (PubMed)
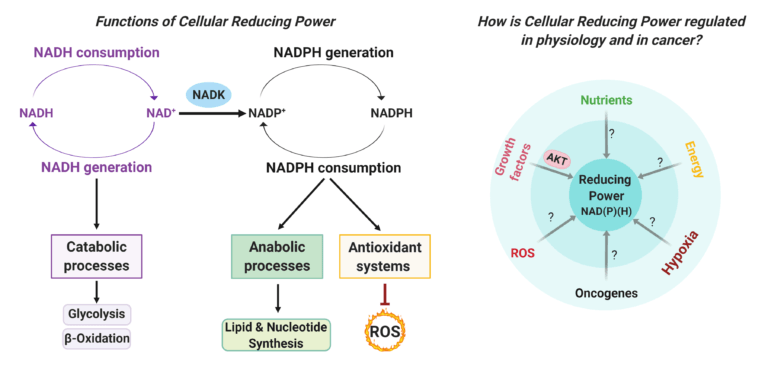
Hoxhaj, G., Ben-Sahra, I., Lockwood, S.E., Timson, R.C., Byles, V., Henning, G.T., Gao, P., Selfors, L.M., Asara, J.M., and B.D. Manning. (2019). Direct stimulation of NADP+ synthesis through Akt-mediated phosphorylation of NAD kinase. Science 363, 1088-1092. (PubMed)
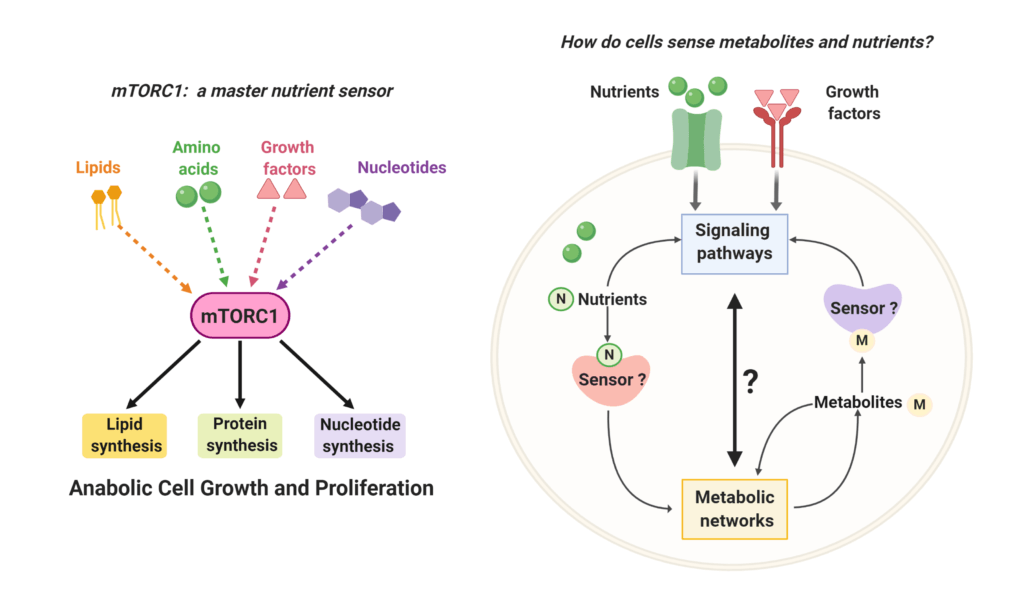
Hoxhaj, G., Hallett, J.H., Timson, R., Ilagan, E., Asara, J.M., Ben-Sahra, I., and B.D. Manning. (2017). The mTORC1 signaling network senses changes in cellular purine nucleotide levels. Cell Reports 21, 1331-1346. (PubMed)
Ben-Sahra, I.,* Hoxhaj, G.,* Ricoult, S.J., Asara, J.M., and B.D. Manning. (2016). mTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle. Science 351, 728-33. (PubMed)
Hoxhaj, G.,# Caddye, E., Najafov, A., Houde, V.P., Johnson, C., Dissanayake, K., Toth, R., Campbell, D.G., Prescott, A.R., and C. MacKintosh.# (2016). The E3 ubiquitin ligase ZNRF2 is a substrate of mTORC1 and regulates its activation by amino acids. eLife pii: e12278. (PubMed)
Hoxhaj, G.,# Najafov, A., Toth, R., Campbell, D.G., Prescott, A.R., and C. MacKintosh.# (2012). ZNRF2 is released from membranes by growth factors and, together with ZNRF1, regulates the Na+/K+ATPase. Journal of Cell Science 125, 4662-75. (PubMed)
*Contributed equally
#Co-corresponding autho