Chiang, J.C., Shang, Z., Rosales, T., Cai, L., Chen, W.M., Cai, F., Vu, H., Minna, J.D., Ni, M., Davis, A.J., Timmerman, R.D., DeBerardinis, R.J.#, and Y. Zhang#. (2025). Lipoylation inhibition enhances radiation control of lung cancer by suppressing homologous recombination DNA damage repair. Science Advances 11:eadt1241. (PubMed)
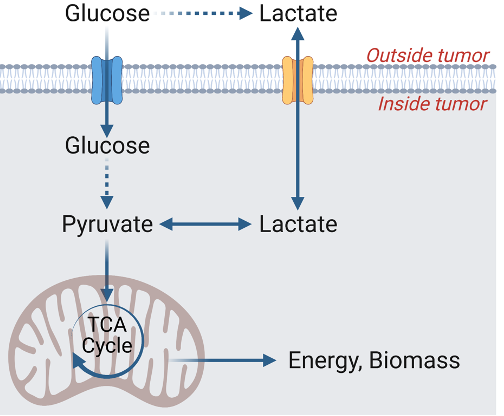
Cai, L., Hammond, N.G., Tasdogan, A., Alsamraae, M., Yang, C., Cameron, R.B., Quan, P., Solmonson, A., Gu, W., Pachnis, P., Kaur, M., Chang, B.K., Zhou, Q., Hensley, C.T., Do, Q.N., Martins Nascentes Melo, L., Ubellacker, J.M., Kaushik, A., Clare, M.G., Alcazar, I.N., Kurylowicz, K., Marcuccilli, J.D., Allies, G., Kutritz, A., Klode, J., Ramesh, V., Rogers, T.J., Rao, A.D., Crentsil, H.E., Li, H., Brister, F., McDaniel, P., Xu, X., Evers, B.M., Zacharias, L.G., Sudderth, J., Xu, J., Mathews, T.P., Oliver, D., Minna, J.D., Waters, J., Morrison, S.J., Kernstine, K.H., Faubert, B.#, and R.J. DeBerardinis#. (2025). High Glucose Contribution to the TCA Cycle Is a Feature of Aggressive Non-Small Cell Lung Cancer in Patients. Cancer Discovery ePub ahead of print. (PubMed)
Bezwada, D., Perelli, L, Lesner, N.P., Cai, L., Brooks, B., Wu, Z., Vu, H.S., Sondhi, V., Cassidy, D.L., Kasitinon, S., Kelekar, S., Cai, F., Aurora, A.B., Patrick, M., Leach, A., Ghandour, R., Zhang, Y., Do, D., McDaniel, P., Sudderth, J., Dumesnil, D., House, S., Rosales, T., Poole, A.M., Lotan, Y., Woldu, S., Bagrodia, A., Meng, X., Cadeddu, J.A., Mishra, P., Garcia-Bermudez, J., Pedrosa, I., Kapur, P., Courtney, K.D., Malloy, C.R., Genovese, G., Margulis, V. and R.J. DeBerardinis. (2024). Mitochondrial complex I promotes kidney cancer metastasis. Nature 633, 923-31. (PubMed)
Wu, Z., Bezwada, D., Cai, F., Harris, R.C., Ko, B., Sondhi, V., Pan, C., Vu, H.S., Nguyen, P.T., Faubert, B., Cai, L., Chen, H., Martin-Sandoval, M., Do, D., Gu, W., Zhang, Y., Zhang, Y., Brooks, B., Kelekar, S., Zacharias, L.G., Oaxaca, K.C., Patricio, J.S., Mathews, T.P., Garcia-Bermudez, J., Ni, M., and R.J. DeBerardinis. (2024). Electron transport chain inhibition increases cellular dependence on puring transport and salvage. Cell Metabolism 36, 1504-20. (PubMed)
Cai, F., Bezwada, D., Cai, L., Mahar, R., Wu, Z., Chang, M.C., Pachnis, P., Yang, C., Kelekar, S., Gu, W., Brooks, B., Ko, B., Vu, H.S., Mathews, T.P., Zacharias, L.G., Martin-Sandoval, M., Do, D., Oaxaca, K.C., Jin, E.S., Margulis, V., Malloy, C.R., Merritt, M.E., and R.J. DeBerardinis. (2023). Comprehensive isotopomer analysis of glutamate and aspartate in small tissue samples. Cell Metabolism 35, 1830-184. (PubMed)
Kaushik, A.K., Tarangelo, A., Boroughs, L.K., Ragavan, M., Zhang, Y., Wu, C.Y., Li, X., Ahumada, K., Chiang, J.C., Tcheuyap, V.T., Saatchi, F., Do, Q.N., Yong, C., Rosales, T., Stevens, C., Rao, A.D., Faubert, B., Pachnis, P., Zacharias, L.G., Vu, H., Cai, F., Mathews, T.P., Genovese, G., Slusher, B.S., Kapur, P., Sun, X., Merritt, M., Brugarolas, J, and R.J. DeBerardinis. (2022). In vivo characterization of glutamine metabolism identifies therapeutic targets in clear cell renal cell carcinoma. Science Advances 8:eabp8293. (PubMed)
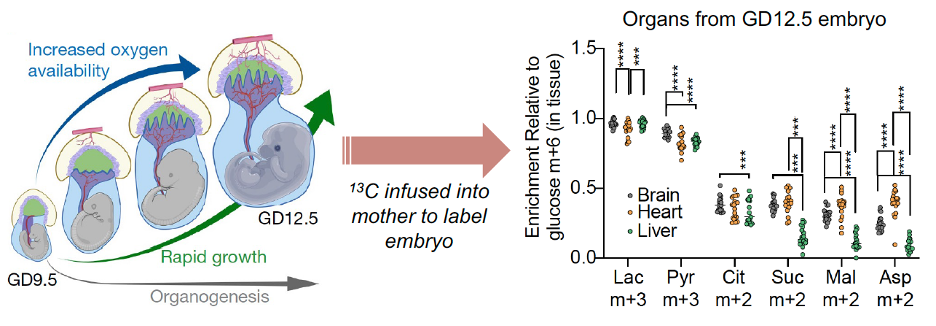
Solmonson, A., Faubert, B., Gu, W., Rao, A., Cowdin, M.A., Mendez-Monte, I., Kelekar, S., Rogers, T.J., Pan, C., Guevara, G., Tarangelo, A., Zacharias, L.G., Martin-Sandoval, M.S., Do, D., Pachnis, P., Dumesnil, D., Mathews, T., Tasdogan, A., Pham, A., Cai, L., Zhao, Z., Ni, M., Cleaver, O., Sadek, H.A., Morrison, S.J., and R.J. DeBerardinis. (2022). Compartmentalized metabolism supports midgestation mammalian development. Nature 604, 349-353. (PubMed)
Pachnis, P., Wu, Z., Faubert, B., Tasdogan, A., Gu, W., Shelton, S., Solmonson, A., Rao, A.D, Kaushik, A.K., Rogers, T.J., Ubellacker, J.M., LaVigne, C.A., Yang, C., Ko, B., Ramesh, V., Sudderth, J., Zacharias, L.G., Martin-Sandoval, M.S., Do, D., Mathews, T.P., Zhao, Z., Mishra, P., Morrison, S.J., and R.J. DeBerardinis. (2022). In vivo isotope tracing reveals a requirement for the electron transport chain in glucose and glutamine metabolism by tumors. Science Advances 8:eabn9550. (PubMed)
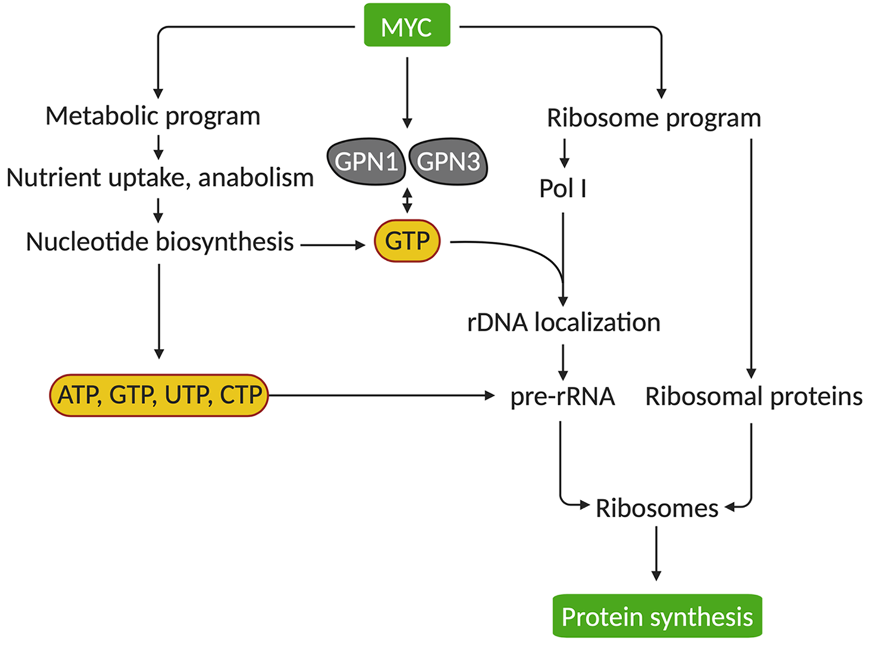
Huang, F., Huffman, K.E., Wang, Z., Wang, X., Cai, F., Yang, C., Cai, L., Shih, T., Zacharias, L.G., Chung, A., Yang, Q., Chalishazar, M.D., Ireland, A.S., Stewart, C.A., Cargill, K., Girard, L., Liu, Y., Ni, M., Xu, J., Wu, X., Zhu, H., Drapkin, B., Oliver, T.G., Byers, L.G., Gazdar, A.F., Minna, J.D., and R.J. DeBerardinis. (2021). Guanosine triphosphate couples oncogenic MYC’s metabolic and ribosome programs. Journal of Clinical Investigation 131:e139929. (PubMed)
Tasdogan, A., Faubert, B., Ramesh, V., Ubellacker, J.M., Shen, B., Solmonson, A., Murphy, M.M., Gu, Z., Gu, W., Martin, M., Kasitinon, S.Y., Vandergriff, T., Mathews, T.P., Zhao, Z., Schadendorf, D., DeBerardinis, R.J., and S.J. Morrison. (2020). Metabolic heterogeneity confers differences in melanoma metastatic potential. Nature 577, 115-120. (PubMed)
Chen, P.H., Cai, L., Huffman, K., Yang, C., Kim, J., Faubert, B., Boroughs, L., Ko, B., Sudderth, J., McMillan, E.A., Girard, L., Chen, D., Peyton, M., Shields, M.D., Yao, B., Shames, D.S., Kim, H.S., Timmons, B., Sekine, I., Britt, R., Weber, S., Byers, L.A., Heymach, J.V., Chen, J., White, M.A., Minna, J.D., Xiao, G., and R.J. DeBerardinis. (2019). Metabolic Diversity in Human Non-Small Cell Lung Cancer Cells. Molecular Cell 76, 838-851. (PubMed)
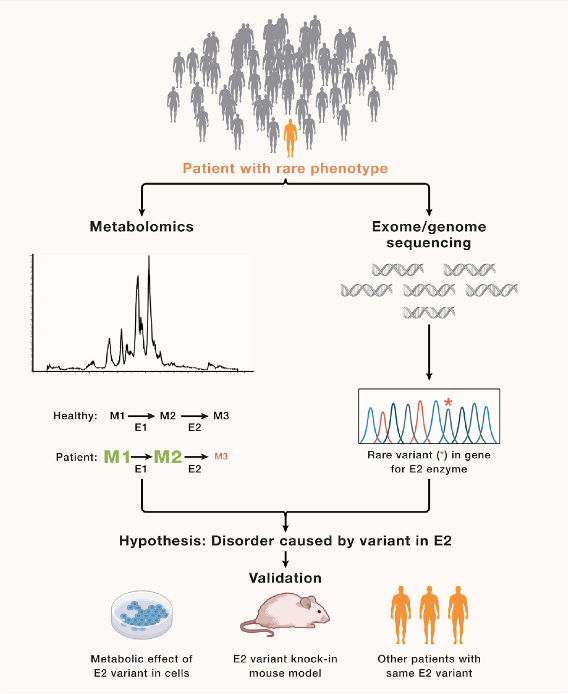
Ni, M., Solmonson, A., Pan, C., Yang, C., Li, D., Notzon, A., Cai, L., Guevara, G., Zacharias, L.G., Faubert, B., Vu, H.S., Jiang, L., Ko, B., Morales, N.M., Pei, J., Vale, G., Rakheja, D., Grishin, N.V., McDonald, J.G., Gotway, G.K., McNutt, M.C., Pascual, J.M., and R.J. DeBerardinis. (2019) Functional Assessment of Lipoyltransferase- Deficiency in Cells, Mice, and Humans. Cell Reports 27, 1376-1386. (PubMed)
Huang, F., Ni, M., Chalishazar, M.D., Huffman, K.E., Kim, J., Cai, L., Shi, X., Cai, F., Zacharias, L.G., Ireland, A.S., Li, K., Gu, W., Kaushik, A.K., Liu, X., Gazdar, A.F., Oliver, T.G., Minna, J.D., Hu, Z., and R.J. DeBerardinis. (2018). Inosine Monophosphate Dehydrogenase Dependence in a Subset of Small Cell Lung Cancers. Cell Metabolism 28, 369-382. (PubMed)
Faubert, B., Li, K.Y., Cai, L., Hensley, C.T., Kim, J., Zacharias, L.G., Yang, C., Do, Q.N., Doucette, S., Burguete, D., Li, H., Huet, G., Yuan, Q., Wigal, T., Butt, Y., Ni, M., Torrealba, J., Oliver, D., Lenkinski, R.E., Malloy, C.R., Wachsmann, J.W, Young, J.D., Kernstine, K., and R.J. DeBerardinis. (2017) Lactate Metabolism in Human Lung Tumors. Cell 171, 358-371. (PubMed)
Kim, J., Hu, Z., Cai, L., Li, K., Choi, E., Faubert, B., Bezwada, D., Rodriguez-Canales, J., Villalobos, P., Lin, Y-F., Ni, M., Huffman, K.E., Girard, L., Byers, L.A., Unsal-Kacmaz, K., Peña, C.G., Heymach, J.V., Wauters, E., Vansteenkiste, J., Castrillon, D.H., Chen, B.P.C., Wistuba, I,, Lambrechts, D., Xu, J., Minna, J.D., and R.J. DeBerardinis. (2017). CPS1 maintains pyrimidine pools and DNA synthesis is KRAS/LKB1-mutant lung cancer cells. Nature 546, 168-172. (PubMed)
Hensley, C.T., Faubert, B., Yuan, Q., Lev-Cohain, N., Jin, E., Kim, J., Jiang, L., Ko, B., Skelton, R., Loudat, L., et al. (2016). Metabolic heterogeneity in human lung tumors. Cell 164, 681–694. (PubMed)
Jiang, L., Shestov, A.A., Swain, P., Yang, C., Parker, S.J., Wang, Q.A., Terada, L.S., Adams, N.D., McCabe, M.T., Pietrak, B., et al. (2016). Reductive carboxylation supports redox homeostasis during anchorage-independent growth. Nature 532, 255–258. (PubMed)
#Co-corresponding authors