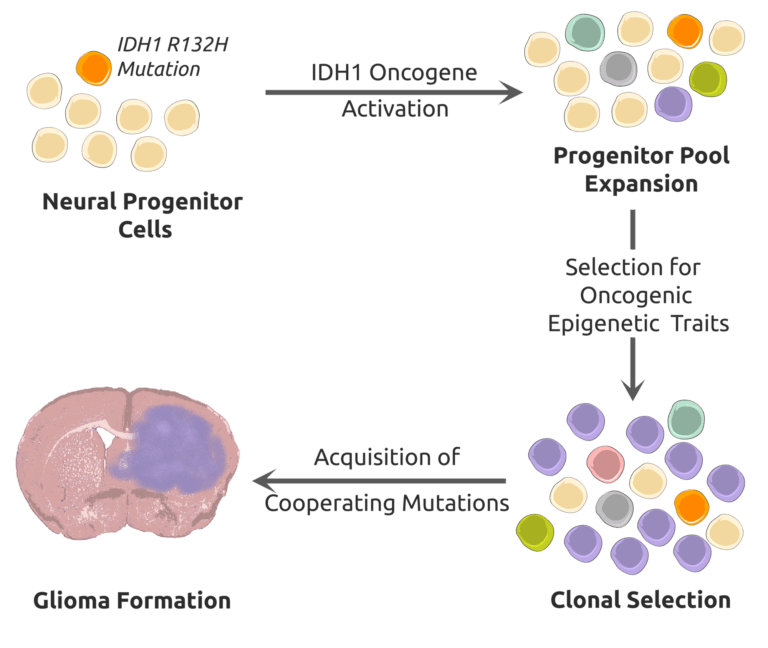
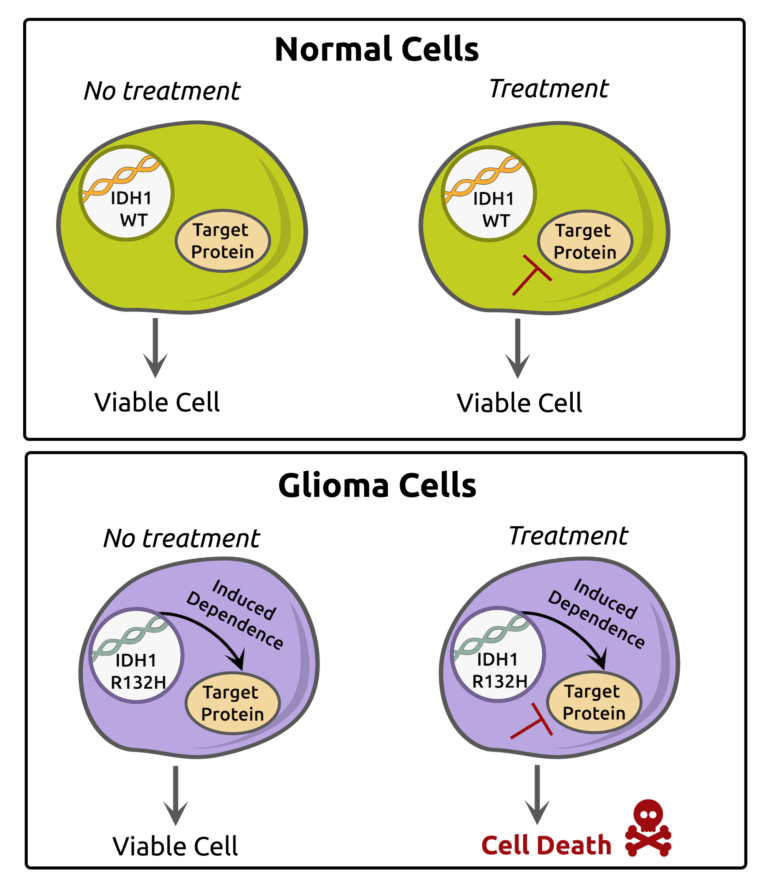
Wu, M.J., Kondo, H., Kammula, A.V., Shi, L., Xiao, Y., Dhiab, S., Xu, Q., Slater, C.J., Avila, O.I., Merritt, J., Kato, H., Kattel, P., Sussman, J., Gritti, I., Eccleston, J., Sun, Y., Cho, H.M., Olander, K., Katsuda, T., Shi, D.D., Savani, M.R., Smith, B.C., Cleary, J.M., Mostoslavsky, R., Vijay, V., Kitagawa, Y., Wakimoto, H., Jenkins, R.W., Yates, K.B., Paik, J., Tassinari, A., Saatcioglu, D.H., Tron, A.E., Haas, W., Cahill, D.P., McBrayer, S.K., Manguso, R.T., and N. Bardeesy. (2024). Mutant IDH1 Inhibition Induces dsDNA Sensing to Activate Tumor Immunity. Science 385:eadl6173. (PubMed)
Shi, D.D., Lee, J.H., Wang, A.C., Khanal, J., Gao, W., Kaelin, W.G. Jr., and S.K. McBrayer. (2023). Protocol to establish a genetically engineered mouse model of IDH1-mutant astrocytoma. STAR Protocols 4:102281. (PubMed)
Nguyen, T.P., Wang, W., Sternisha, A.C., Corley, C.D., Wang, H.Y.L., Wang, X., Ortiz, F., Lim, S.K., Abdullah, K.G., Parada, L.F., Williams, N.S., McBrayer, S.K., McDonald, J.G., De Brabander, J.K., and D. Nijhawan. (2023). Selective and Brain-penetrant Lanosterol Synthase Inhibitors Target Glioma Stem-like Cells by Inducing 24(S),25-epoxycholesterol Production. Cell Chemical Biology 30, 214-229. (PubMed)
Miller, J.J., Gonzalez Castro, L.N., McBrayer, S.K., Weller, M., Cloughesy, T., Portnow, J., Andronesi, O., Barnholtz-Sloan, J.S., Baumert, B.G., Berger, M.S., Bi, W.L., Bindra, R., Cahill, D.P., Chang, S.M., Costello, J.F., Horbinski, C., Huang, R.Y., Jenkins, R.B., Ligon, K.L., Mellinghoff, I.K., Nabors, L.B., Platten, M., Reardon, D.A., Shi, D.D., Schiff, D., Wick, W., Yan, H., von Deimling, A., van den Bent, M., Kaelin, W.G., and P.Y. Wen. (2022). Isocitrate dehydrogenase (IDH) mutant gliomas: A Society for Neuro-Oncology (SNO) consensus review on diagnosis, management, and future directions. Neuro Oncology 25, 4-25. (PubMed)
Notarangelo, G., Spinelli, J.B., Perez, E.M., Baker, G.J., Kurmi, K., Elia, I., Stopka, S.A., Baquer, G., Lin, J.R., Golby, A..J, Joshi, S., Baron, H.F., Drijvers, J.M., Georgiev, P., Ringel, A.E., Zaganjor, E., McBrayer, S.K., Sorger, P.K., Sharpe, A.H., Wucherpfennig, K.W., Santagata, S., Agar, N.Y.R., Suvà, M.L., and M.C. Haigis. (2022). Oncometabolite d-2HG alters T cell metabolism to impair CD8+ T cell function. Science 377, 1519-1529. (PubMed)
Pal, S., Kaplan, J.P., Nguyen, H., Stopka, S.A., Savani, M.R., Regan, M.S., Nguyen, Q.D., Jones, K.L., Moreau, L.A., Peng, J., Dipiazza, M.G., Perciaccante, A.J., Zhu, X., Hunsel, B.R., Liu, K.X., Alexandrescu, S., Drissi, R., Filbin, M.G., McBrayer, S.K., Agar, N.Y.R., Chowdhury, D., and D.A. Haas-Kogan. (2022). A druggable addiction to de novo pyrimidine biosynthesis in diffuse midline glioma. Cancer Cell 40, 957-972. (PubMed)
Shi, D.D., Savani, M.R., Levitt, M.M., Wang, A.C., Endress, J.E., Bird, C.E., Buehler, J., Stopka, S.A., Regan, M.S., Lin, Y.F., Puliyappadamba, V.T., Gao, W., Khanal, J., Evans, L., Lee, J.H., Guo, L., Xiao, Y., Xu, M., Huang, B., Jennings, R.B., Bonal, D.M., Martin-Sandoval, M.S., Dang, T., Gattie, L.C., Cameron, A.B., Lee, S., Asara, J.M., Kornblum, H.I., Mak, T.W., Looper, R.E., Nguyen, Q.D., Signoretti, S., Gradl, S., Sutter, A., Jeffers, M., Janzer, A., Lehrman, M.A., Zacharias, L.G., Mathews, T.P., Losman, J.A., Richardson, T.E., Cahill, D.P., DeBerardinis, R.J., Ligon, K.L., Xu, L., Ly, P., Agar, N.Y.R., Abdullah, K.G., Harris, I.S., Kaelin, W.G. Jr, and S.K. McBrayer. (2022). De novo pyrimidine synthesis is a targetable vulnerability in IDH mutant glioma. Cancer Cell 40, 939-956. (PubMed)
Guo, G., Gong, K., Beckley, N., Zhang, Y., Yang, X., Chkheidze, R., Hatanpaa, K.J., Garzon-Muvdi, T., Koduru, P., Nayab, A., Jenks, J., Sathe, A.A., Liu, Y., Xing, C., Wu, S.Y., Chiang, C.M., Mukherjee, B., Burma, S., Wohlfeld, B., Patel, T., Mickey, B., Abdullah, K., Youssef, M., Pan, E., Gerber, D.E., Tian, S., Sarkaria, J.N., McBrayer, S.K., Zhao, D., and A.A. Habib. (2022). EGFR ligand shifts the role of EGFR from oncogene to tumour suppressor in EGFR-amplified glioblastoma by suppressing invasion through BIN3 upregulation. Nature Cell Biology 24, 1291-1305. (PubMed)
Abdullah, K.G., Bird, C.E., Buehler, J.D., Gattie, L.C., Savani, M.R., Sternisha, A.C., Xiao, Y., Levitt, M.M., Hicks, W.H., Li, W., Ramirez, D.M.O., Patel, T., Garzon-Muvdi, T., Barnett, S., Zhang, G., Ashley, D.M., Hatanpaa, K.J., Richardson, T.E., and S.K. McBrayer. (2022). Establishment of patient-derived organoid models of lower-grade glioma. Neuro Oncology 24, 612-623. (PubMed)
Koduri, V., McBrayer, S.K., Liberzon, E., Wang, A.C., Briggs, K.J., Cho, H., and W.G. Kaelin. (2019). Peptidic Degron for IMiD-Induced Degradation of Heterologous Proteins. PNAS 116, 2539-2544. (PubMed)
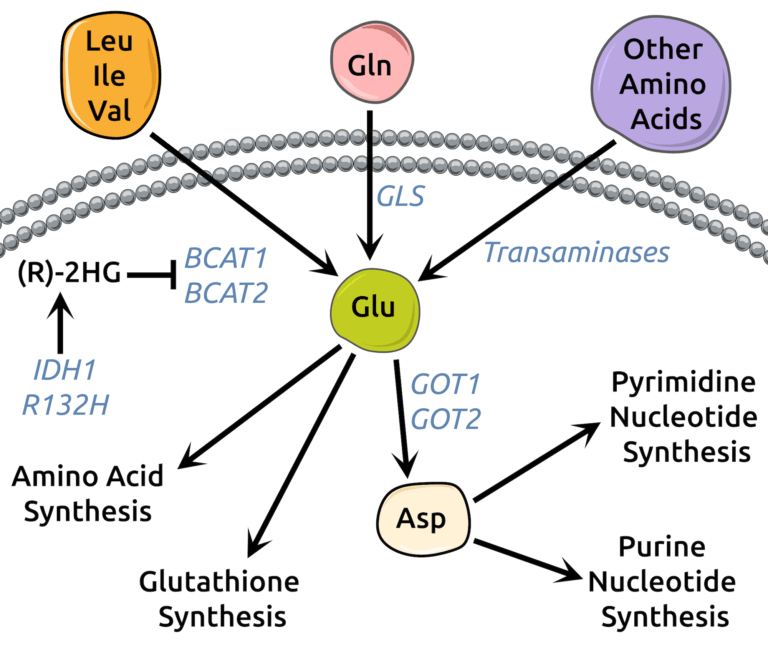
McBrayer, S.K., Mayers, J.R., DiNatale, G.J., Shi, D.D., Khanal, J., Chakraborty, A.A., Sarosiek, K.A., Briggs, K.J., Robbins, A.K., Sewastianik, T., Shareef, S.J., Olenchock, B.A., Parker, S.J., Tateishi, K., Spinelli, J.B., Islam, M., Haigis, M.C., Looper, R.E., Ligon, K.L., Bernstein, B.E., Carrasco, R.D., Cahill, D.P., Asara, J.M., Metallo, C.M., Yennawar, N.H., Vander-Heiden, M.G., and W.G. Kaelin. (2018). Transaminase Inhibition by 2-Hydroxyglutarate Impairs Glutamate Biosynthesis and Redox Homeostasis in Glioma. Cell 175, 101-116. (PubMed)
McBrayer, S.K., Olenchock, B.A., DiNatale, G.J., Shi, D.D., Khanal, J., Jennings, R.B., Novak, J.S., Oser, M.G., Robbins, A.K., Modiste, R., Bonal, D., Moslehi, J., Bronson, R.T., Neuberg, D., Nguyen, Q.D., Signoretti, S., Losman, J.A., and W.G. Kaelin. (2018). Autochthonous Tumors Driven by Rb1 Loss Have an Ongoing Requirement for the RBP2 Histone Demethylase. PNAS 115, E3741-E3748. (PubMed)