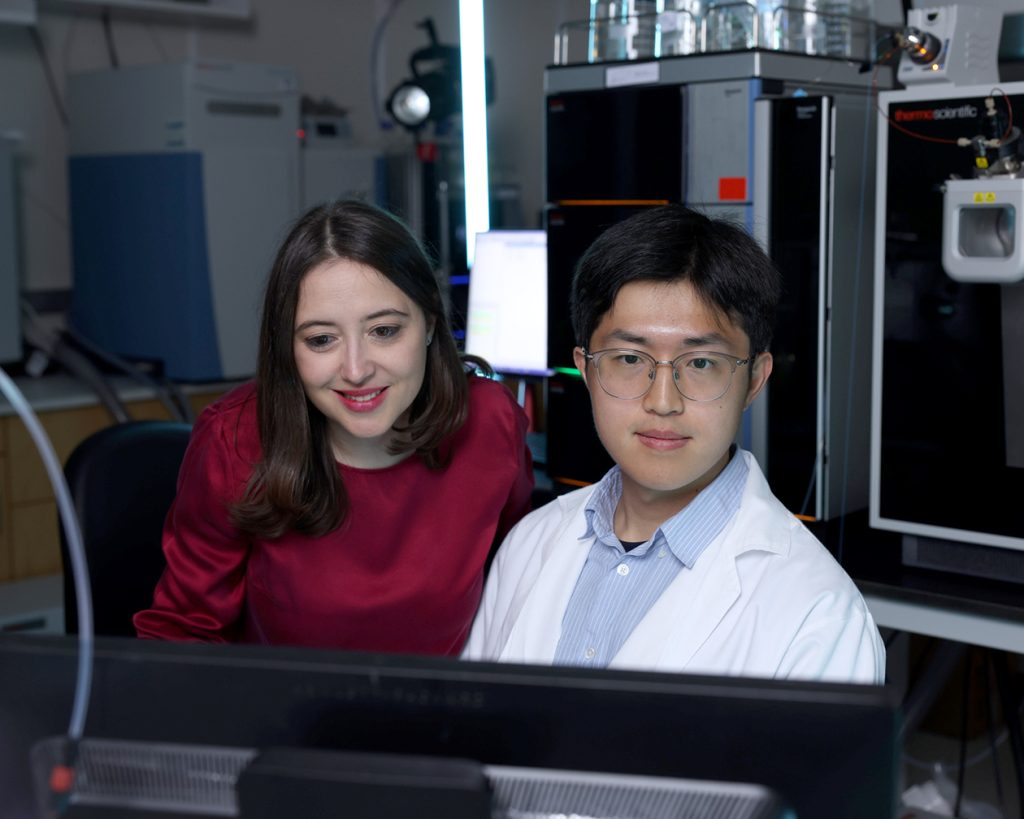
Researchers from the DeBerardinis lab have found a new metabolic liability in chemotherapy-resistant small cell lung cancer (SCLC), one of the most deadly forms of human cancer. In doing so, they also uncovered a fundamental aspect of how cancer cells coordinate growth regulated by MYC, one of the most commonly activated oncogenes across many human cancers. Both findings pinpointed molecular components of aggressive tumors that can be targeted by existing drugs.
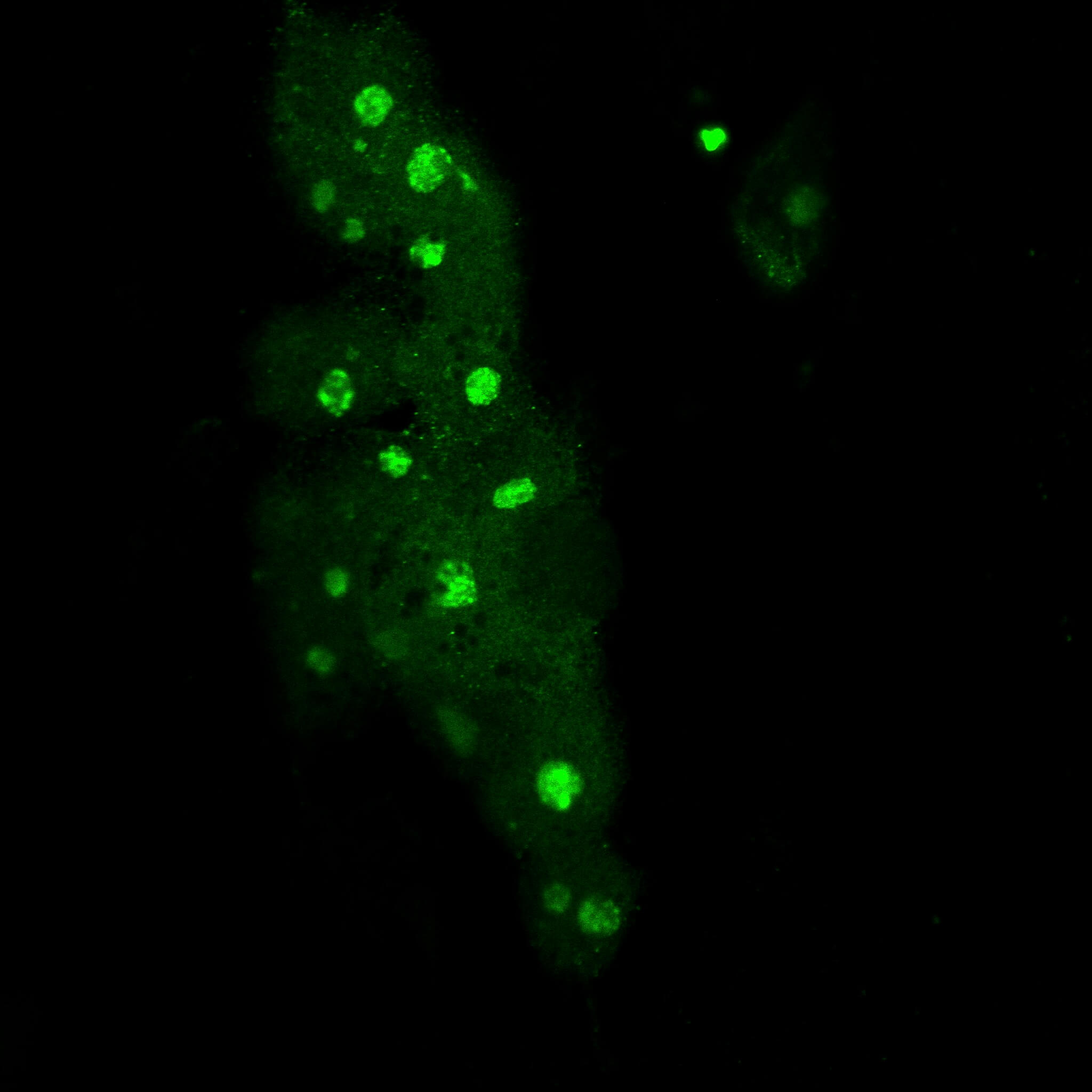
Immunofluorescence MYC-high cells.
“Most patients with SCLC respond to chemotherapy at first, but the tumor quickly becomes drug-resistant, re-grows, and kills the patient within a few months. With a five-year survival rate of six percent, we badly need new treatments, particularly in drug-resistant SCLC,” says Ralph DeBerardinis, M.D., Ph.D., a professor at Children’s Medical Center Research Institute at UT Southwestern (CRI) and a Howard Hughes Medical Institute Investigator.
Previous research from the DeBerardinis lab linked high levels of MYC with high rates of guanosine triphosphate (GTP) synthesis in an aggressive subset of SCLC. In these cancers, MYC activated the genes responsible for encoding an enzyme in the GTP pathway, inosine monophosphate dehydrogenase (IMPDH). Blocking the GTP pathway with mizoribine, an IMPDH inhibitor, resulted in suppressed tumor growth in mice with MYC-expressing SCLC.
Building on this work, the lab compared metabolic liabilities between SCLCs that had not been exposed to chemotherapy and SCLCs that had become chemotherapy resistant. They discovered that chemotherapy-resistant SCLC tumors acquired dependence on the IMPDH pathway and could be treated with IMPDH inhibitors. This addiction to IMPDH resulted from the fact that chemotherapy-resistant tumors tended to activate MYC, which then drove GTP synthesis as it did in tumors with high levels of MYC from the start. The researchers also used genetic models of cancer in mice to show that tumors driven by MYC in other parts of the body, including the liver, are also highly dependent on the GTP pathway and can be treated with IMPDH inhibitors.

Fang Huang, Ph.D.
In the study, published in the Journal of Clinical Investigation, the team also uncovered a new aspect of how MYC drives cell growth. Researchers have known for many years that MYC promotes cellular metabolism and simultaneously activates production of ribosomes, which allow cells to make proteins to grow. The DeBerardinis lab showed that the GTP pathway ties together MYC’s metabolic effects to its effects on protein synthesis. They determined that GTP provides a signal to allow cells to activate ribosome production. When GTP stores became depleted, the cells could not start the process of synthesizing ribosomes even when MYC was present.
“We think that GTP is the key metabolic signal that connects MYC’s metabolic effects to its effects on protein synthesis and cancer cell growth,” says Fang Huang, Ph.D., the postdoctoral fellow in the DeBerardinis lab who led the study and is now a practicing oncologist in China. “If the cell does not have enough GTP, it cannot activate protein synthesis no matter what other metabolic pathways are present.” The team went on to identify specific proteins that detect GTP and turn on the process of ribosome production. Blocking these GTP-sensing proteins also suppressed growth of chemotherapy-resistant SCLC cells.
“Fang’s work in this paper not only helps us understand the mechanism behind activation of the GTP synthesis, but also identifies other possible therapeutic targets beyond the GTP pathway to slow the growth of tumors with high levels of MYC,” says Dr. DeBerardinis.